20.109(S14):DNA cloning (Day4)
Introduction
Welcome back! Last time you performed PCR on the DNA pool you extracted from a gull sample. Specifically, you used primers designed to amplify bacterial 16S rRNA gene fragments from this complex pool. Now you want to systematically analyze individual fragments from this 16S DNA pool, which is aided by a process called cloning.
In traditional cloning, an insert is ligated to a backbone to create a circular piece of DNA called a vector. Typically, the insert and backbone are cut with special proteins called restriction enzymes (more about those in Module 2!) that yield complementary overhangs at the ends of the two pieces of DNA. The enzyme DNA ligase then chemically bonds the insert and backbone together by catalyzing formation of a phosphodiester bond. Ligated vectors are selected in bacteria by including an antibiotic resistance marker on the backbone, and growing the bacteria in said antibiotic. (We’ll say more about that procedure below.) This method is especially suitable for cloning with the purpose of creating a well-defined piece of DNA.

When diverse products must be cloned for subsequent analysis, as in our case, we would like to omit some steps from the above workflow. A few methods exist for cloning PCR products directly. The most popular approach is dubbed TA cloning, and it exploits the fact that polymerases such as Taq add an adenine to the 3’ end of PCR products. This single-base overhang alone can be used in a process very much like traditional cloning. Recall, however, that we used Pfu polymerase precisely for its precision, to improve our chance of successful amplification from a “difficult” sample such as stool. Pfu has proofreading capabilities, meaning that it doesn’t add spurious adenines.
Pieces of DNA that do not share complementary overhangs, that indeed have no overhangs at all but instead have blunt ends, can also be ligated together. There are several mechanisms by which this ligation may be accomplished. The simplest method and the one we will employ is the use of ligase. The enzyme must be highly efficient: blunt-ended DNA is not joined as easily as DNA with overhangs. An example of another method is so-called TOPO cloning, in which the biology of DNA topoisomerase I is exploited. Here the backbone includes two CCCTT sequences at the site where the PCR product will be inserted. Each sequence is cut by topoisomerase and remains bound to it; the two ends can ligate back to each other or to a newly introduced insert sequence. Whatever the method for ligation, the reaction mixture – in our case buffer, ligase, pCR-Blunt vector, and the PCR insert from M1D3 – is then added to bacteria.
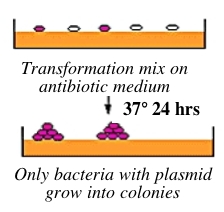
Bacteria can take up foreign DNA in a process called transformation, during which a single plasmid enters a bacterium and, once inside, replicates and expresses the genes it encodes. Most bacteria do not exist in a transformation-ready state, but can be made permeable to foreign DNA -- by chemical or other means -- and are then termed competent. Competent cells are extremely fragile and should be handled gently, specifically kept cold and not vortexed. The transformation procedure is efficient enough for most lab purposes, with efficiencies as high as 109 transformed cells per microgram of DNA, but it is important to realize that even with high efficiency cells only 1 DNA molecule in about 10,000 is successfully transformed.

After transformation, the bacteria are plated on an antibiotic-containing agar medium. In your case, the pCR-Blunt vector has a kanamycin resistance gene, and thus only bacteria that took up the vector can grow on kanamycin-containing plates, while untransformed cells will die before they can form a colony. Notice that a vector-containing bacterium will grow whether or not that vector contains a PCR product insert.
We would rather not waste our time analyzing colonies that contain pure backbone DNA! Thus, besides the KanR gene, pCR-Blunt contains a ccdB gene, which produces a protein that is toxic to E coli; successful ligation of an insert interrupts the ccdB sequence, allowing bacteria containing this plasmid to survive. The ccdB gene is fused to lacZalpha and expressed under the Plac promoter, which can be turned on by the small molecule inducer IPTG, or is constitutively on in cells lacking lacI (the lac repressor gene). The One Shot TOP10 cells have just this genotype; how convenient! The cloning vector is shown with its key features at right.
So today you’ll ligate and transform your PCR product pool, but only after visualizing that everything looks as expected on an agarose gel. Next time you will isolate DNA clones from individual colonies and send them off for sequencing. Then we’ll be just one step away from knowing what bacteria (a sampling of them, in any case!) are present in each gull sample.
Protocols
Part 1: Gel electrophoresis of PCR products
You will use a 1.5% agarose gel to run your three PCRs from last time, as well as a reference lane of molecular weight markers (also called a DNA ladder).
- Add 4 μL of loading dye to each of three eppendorf tubes. No need to change tips in between.
- Loading dye contains xylene cyanol as a tracking dye to follow the progress of the electrophoresis (so you don’t run the smallest fragments off the end of your gel!) as well as glycerol to help the samples sink into the well.
- Now add 20 μL of each reaction from last time (your sample, your partner's sample, and the no template control) to individual eppendorf tubes and label them.
- Flick the eppendorf tubes to mix the contents, then quick spin them in the microfuge to bring the contents of the tubes to the bottom.
- Remember: to quick-spin, hold down the "short" button on your centrifuge for 3-5 seconds, then release.
- Load the gel according to the table below. Up to 4 groups will share each gel: 2 groups per lane.
- To load your samples, draw 20 μL into the tip of your P20. Lower the tip below the surface of the buffer and directly over the well. You risk puncturing the bottom of the well if you lower the tip too far into the well itself (puncturing well = bad!). Slowly expel your sample into the well. Do not release the pipet plunger until after you have removed the tip from the gel box (or you'll draw your sample back into the tip!).
- Once all the samples have been loaded, we will attach the gel box to the power supply and run at 110 V for 45 minutes.
- Later you will be shown how to photograph your gel and, if necessary, to excise the relevant band of DNA. Begin by anticipating where you expect to see your sample band relative to the bands of the DNA ladder, described here.

| Lane | Sample (20 μL) | Lane | Sample (20 μL) |
|---|---|---|---|
| 1 | DNA ladder (load 10 μL) | 6 | DNA ladder (load 10 μL) |
| 2 | Group 1, NTC | 7 | Group 2, NTC |
| 3 | Group 1, sample A | 8 | Group 2, sample A |
| 4 | Group 1, sample B | 9 | Group 2, sample B |
| 5 | BLANK | 10 | BLANK |
Part 1B: OPTIONAL -- Purify 16S PCR band from gel
In many types of cloning, PCR products are purified prior to ligation. For the cloning approach that we are taking, the manufacturer recommends using PCR products directly. Indeed, in pilot experiments, we found that cloning was at least as successful with direct PCR products as with gel-purified PCR products. However, if your PCR resulted in multiple bright bands on your agarose gel, then you will need to isolate the 16S band before proceeding.
In gel purification, a DNA band is melted, then isolated on a silica (SiO2) column similar to the one you used last time. Salt concentration and pH effects, along with ethanol precipitation, will alternately allow for binding and eluting the DNA while washing away contaminants. The protocol is linked separately here to avoid clutter on this page.
Part 2: Ligate purified product to backbone
Will they each clone this time (pricy) or still choose the brighter band? Unlikely to be different because not distributed across two groups this time. Also… have them practice a backbone/insert ligation calc for homework? Nice skill to do/have.
- First, choose whichever PCR yielded a brighter band on the gel (assuming no non-specific products in either), and perform one ligation and transformation together. Ultimately, you will each prepare your own plate of cells. Please save both extra PCR reactions and leftover ligation mix today. These will be stored frozen in case of later needs.
- For example, if Shannon and Agi both worked with gull sample 842, and Shannon's band was brighter, then Shannon and Agi would perform the procedure below with Shannon's PCR, but they wouldn't throw away Agi's PCR either.
- Prepare a clearly labeled eppendorf tube for the cloning reaction.
- Our goal is to combine 25 ng of vector with insert in a 10:1 molar ratio, using no more than 5 μL of insert.
- In the following order, combine water for a final reaction volume of 10 μL, 2 μL reaction buffer, up to 5 μL DNA, 1 μL of pCR-Blunt vector, and 1 μL of ExpressLink T4 DNA ligase.
- Incubate at room temperature for 15 min. Meanwhile, obtain an aliquot of competent cells from the teaching faculty and let them slowly thaw on ice -- takes about 5-10 min. Label the side of the tube (and also the top if you like).
- When both cells and reaction are ready, add 2 μL of the reaction to the cells. Do not pipet up-and-down more than once! Instead, gently mix the cells and DNA by twirling the tube in your fingers.
- Hold the tube near the top, so that the cells at the bottom stay cold.
- Incubate the mixture on ice for 30 min.
- Place the tubes in the 42 °C heat block for exactly 45 seconds, and transfer immediately immediately to ice for 2 min more.
- Add 250 μL of warm LB to each sample; do not pipet to mix.
- Place the tubes on the nutator in the 37 °C incubator, and rock them for about 1 hr.
- You may want to put a colored sticky label on the tube so you can quickly identify it later.
- Shortly after starting your incubation, label and pre-warm two LB-KAN plates from the incubator (team/day/initials/sampleID/date).
Part 3: Prepare tubes for liquid O/N cultures
You will make your teaching faculty very happy if you contribute to their preparatory work.
- Please label 8 large glass test tubes with your team color and sample ID, per sample.
- Mix 25 mL LB with 25 μL of kanamycin.
- Using a serological pipet, aliquot 2.5 mL of LB+Kan per tube. These will be used to set up liquid overnight cultures from your 8 colonies for next time.
Part 4: Transform ligated product into cloning strain
- Pick up an ethanol jar, spreader, lighter, and alcohol burner when you are almost ready to begin. Don't forget to wear your safety glasses!
- Plate 100 μL of each transformation mix onto an LB-Kan plate. After dipping the glass spreader in the ethanol jar, you should pass it through the flame of the alcohol burner just long enough to ignite the ethanol. After letting the ethanol burn off, the spreader may still be very hot, and it is advisable to tap it gently on a portion of the agar plate without cells in order to equilibrate it with the agar (if it sizzles, it's way too hot). Once the plates are ready, place them in the 37°C incubator overnight; be sure that each is labeled with your team color, gull sample ID, and initials. One of the teaching faculty will remove them from the incubator and set up liquid cultures for you to use next time.
For next time
Move schematic/context para earlier, and introduction later??
Parts 2, 3, and 4 of this assignment are also due on Stellar.
- Prepare the overview schematic of your experimental approach that will be included in the Introduction or (more likely) at the beginning of the Results.
- Recall that our collaboration and integrity guidelines state that your text and figures must be your own unless otherwise specified. Thus, you should all make your own schematic rather than modifying one from class slides or the wiki. Keep in mind that you are being evaluated not on advanced PowerPoint skills, but instead on showing that you understand the purpose and major steps of the experiment and can convey them.
- Write the opening context-setting paragraph of your Results section, in which you introduce the system you are working with and your immediate investigative goal.
- This paragraph might be somewhat longer or shorter depending on the level of detail in your Introduction section, but it cannot be omitted entirely.
- Draft an outline of the rest of the results to indicate the approximate high-level content and its order. You don't need to state the actual results (in fact, you don't have them all yet!), just each type of result that will be discussed in about a sentence each. Essentially, what order will your figures/tables/data be in and what will they consist of? Also include sub-section titles in this draft outline.
- Prepare a figure depicting your PCR gel results (from today) with an appropriate caption. Also write the portion of your Results (likely one rather short paragraph) describing this experiment.
Reagent list
- Zero Blunt PCR Cloning kit from Life Technologies
- Includes One Shot TOP10 chemically competent cells
- Luria-Bertani broth from Teknova
- 1% Tryptone
- 0.5 % Yeast extract
- 1 % NaCl
- Optional: Qiagen QIAquick gel extraction kit
- silica spin columns
- QG and PE buffers
- Isopropanol
- Kanamycin: 25 mg/mL, aqueous, sterile-filtered
- LB+KAN plates
- LB with 2% agar and 25 μg/ml Ampicillin
Next Day: DNA sequencing Previous Day: 16S PCR and paper discussion
