20.109(S14):DNA cloning (Day4): Difference between revisions
(New page: {{Template:20.109(S14)}} <div style="padding: 10px; width: 640px; border: 5px solid #FF6600;"> ==Introduction== ==Protocols== ==For next time== ==Reagent list== write something here ...) |
|||
| Line 4: | Line 4: | ||
==Introduction== | ==Introduction== | ||
Welcome back! Last time you performed PCR on the DNA pool you extracted from a gull sample. Specifically, you used primers designed to amplify bacterial 16S rRNA gene fragments from this complex pool. Now you want to systematically analyze individual fragments from this 16S DNA pool, which is aided by a process called cloning. | |||
In traditional cloning, an insert is ligated to a backbone to create a circular piece of DNA called a vector. Typically, the insert and backbone are cut with special proteins called restriction enzymes (more about those in Module 2!) that yield complementary overhangs at the ends of the two pieces of DNA. The enzyme DNA ligase then chemically bonds the insert and backbone together by catalyzing formation of a phosphodiester bond. Ligated vectors are selected in bacteria by including an antibiotic resistance marker on the backbone, and growing the bacteria in said antibiotic. (We’ll say more about that procedure below.) This method is especially suitable for cloning with the purpose of creating a well-defined piece of DNA. | |||
[[Image:20109_cloning-traditional.jpg|thumb|center|550px|'''Cloning overview: traditional approach.''' An insert, such as a PCR product, and a backbone, such as a commercially available vector, are depicted in black and blue, respectively. These are digested with the same restriction enzymes (dubbed "X" and "E") so as to have complementary overhangs, and then usually gel-purified. Finally, backbone and insert are joined together to create a new vector using DNA ligase. ''Note: Figure modified from Prof. Bevin Engelward.'']] | |||
<br style="clear:both;"/> | |||
When diverse products must be cloned for subsequent analysis, as in our case, we would like to omit some steps from the above workflow. A few methods exist for cloning PCR products directly. The most popular approach is dubbed TA cloning, and it exploits the fact that polymerases such as ''Taq'' add an adenine to the 3’ end of PCR products. This single-base overhang alone can be used in a process very much like traditional cloning. Recall, however, that we used ''Pfu'' polymerase precisely for its precision, to improve our chance of successful amplification from a “difficult” sample such as stool. ''Pfu'' has proofreading capabilities, meaning that it doesn’t add spurious adenines. | |||
Pieces of DNA that do not share complementary overhangs, that indeed have no overhangs at all but instead have blunt ends, can also be ligated together. There are several mechanisms by which this ligation may be accomplished…. One is blah. We will simply do bleu. | |||
Here the biology of DNA topoisomerase I is exploited. The backbone includes a CCCTT sequence, which is cut by the topoisomerase, resulting in so-called two vector arms. The vector arms remain bound to the topoisomerase, and can ligate back to each other or to a newly introduced insert sequence. The reaction mixture – buffer, vector arms, and insert (in our case the PCR from M1D3) – is then added to bacteria. | |||
[[Image:Be109transformation.jpg|thumb|right|225px|'''Bacterial transformation.''']] | |||
ADD BACK SOME DETAILS | |||
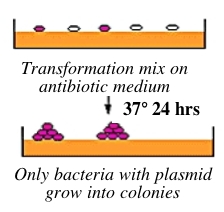
Bacteria can take up foreign DNA in a process called transformation, during which a single plasmid enters a bacterium and, once inside, replicates and expresses the genes it encodes. Most bacteria do not exist in a transformation-ready state, but can be made permeable to foreign DNA -- by chemical or other means -- and are then termed competent. After transformation, the bacteria are plated on an antibiotic-containing agar medium. In your case, the vector arms have an ampicillin resistance gene, and thus only bacteria that took up the vector can grow on ampicillin-containing plates, while untransformed cells will die before they can form a colony. Notice that a vector-containing bacterium will grow whether or not that vector contains a PCR product insert. | |||
<br style="clear:both;"/> | |||
We would rather not waste our time analyzing colonies that contain pure vector DNA! Thus, the reaction mixture is added to a particular strain of bacteria that has a mutation in… | |||
The cloning process as a whole is summarized in the figure below. | |||
[[Image:20109_StrataCloneBlunt-Fig1.jpg|thumb|center|600px|'''Cloning overview: blunt-end cloning.''' Figure very slightly modified from StrataClone Blunt manual, original by Agilent Technologies.]] | |||
So today you’ll ligate and transform your PCR product pool, but only after visualizing that everything looks as expected on an agarose gel. Next time you will isolate DNA clones from individual colonies and send them off for sequencing. Then we’ll be just one step away from knowing what bacteria (a sampling of them, in any case!) are present in each gull sample. | |||
==Protocols== | ==Protocols== | ||
Revision as of 09:13, 3 January 2014
Introduction
Welcome back! Last time you performed PCR on the DNA pool you extracted from a gull sample. Specifically, you used primers designed to amplify bacterial 16S rRNA gene fragments from this complex pool. Now you want to systematically analyze individual fragments from this 16S DNA pool, which is aided by a process called cloning.
In traditional cloning, an insert is ligated to a backbone to create a circular piece of DNA called a vector. Typically, the insert and backbone are cut with special proteins called restriction enzymes (more about those in Module 2!) that yield complementary overhangs at the ends of the two pieces of DNA. The enzyme DNA ligase then chemically bonds the insert and backbone together by catalyzing formation of a phosphodiester bond. Ligated vectors are selected in bacteria by including an antibiotic resistance marker on the backbone, and growing the bacteria in said antibiotic. (We’ll say more about that procedure below.) This method is especially suitable for cloning with the purpose of creating a well-defined piece of DNA.

When diverse products must be cloned for subsequent analysis, as in our case, we would like to omit some steps from the above workflow. A few methods exist for cloning PCR products directly. The most popular approach is dubbed TA cloning, and it exploits the fact that polymerases such as Taq add an adenine to the 3’ end of PCR products. This single-base overhang alone can be used in a process very much like traditional cloning. Recall, however, that we used Pfu polymerase precisely for its precision, to improve our chance of successful amplification from a “difficult” sample such as stool. Pfu has proofreading capabilities, meaning that it doesn’t add spurious adenines.
Pieces of DNA that do not share complementary overhangs, that indeed have no overhangs at all but instead have blunt ends, can also be ligated together. There are several mechanisms by which this ligation may be accomplished…. One is blah. We will simply do bleu.
Here the biology of DNA topoisomerase I is exploited. The backbone includes a CCCTT sequence, which is cut by the topoisomerase, resulting in so-called two vector arms. The vector arms remain bound to the topoisomerase, and can ligate back to each other or to a newly introduced insert sequence. The reaction mixture – buffer, vector arms, and insert (in our case the PCR from M1D3) – is then added to bacteria.
ADD BACK SOME DETAILS
Bacteria can take up foreign DNA in a process called transformation, during which a single plasmid enters a bacterium and, once inside, replicates and expresses the genes it encodes. Most bacteria do not exist in a transformation-ready state, but can be made permeable to foreign DNA -- by chemical or other means -- and are then termed competent. After transformation, the bacteria are plated on an antibiotic-containing agar medium. In your case, the vector arms have an ampicillin resistance gene, and thus only bacteria that took up the vector can grow on ampicillin-containing plates, while untransformed cells will die before they can form a colony. Notice that a vector-containing bacterium will grow whether or not that vector contains a PCR product insert.
We would rather not waste our time analyzing colonies that contain pure vector DNA! Thus, the reaction mixture is added to a particular strain of bacteria that has a mutation in…
The cloning process as a whole is summarized in the figure below.

So today you’ll ligate and transform your PCR product pool, but only after visualizing that everything looks as expected on an agarose gel. Next time you will isolate DNA clones from individual colonies and send them off for sequencing. Then we’ll be just one step away from knowing what bacteria (a sampling of them, in any case!) are present in each gull sample.
Protocols
For next time
Reagent list
write something here or not accessible to edit
