20.109(S10):Module 1: Difference between revisions
(New page: {{Template:20.109(S10)}} <div style="padding: 10px; width: 640px; border: 5px solid #996600;"> ==Module 1== '''Instructors:''' [http://web.mit.edu/be/people/niles.htm Jacquin Niles] an...) |
|||
| (8 intermediate revisions by 2 users not shown) | |||
| Line 7: | Line 7: | ||
'''Instructors:''' [http://web.mit.edu/be/people/niles.htm Jacquin Niles] and [[User:AgiStachowiak| Agi Stachowiak]] | '''Instructors:''' [http://web.mit.edu/be/people/niles.htm Jacquin Niles] and [[User:AgiStachowiak| Agi Stachowiak]] | ||
'''TA:''' Christina Birch | '''TA:''' [[User:Christina_M_Birch| Christina Birch]] | ||
In this module you will investigate RNA aptamer selection. You may already be familiar with peptides or proteins, such as antibodies, that bind to specific molecules. Short fragments of RNA – called RNA aptamers - can have secondary structures that also allow them to bind a target molecule with good affinity and specificity. Normally, RNA aptamers that bind particular targets are found by screening many candidates at random in a process called SELEX, or systematic evolution of ligands by exponential enrichment; however, predictive computational tools can also be used. In the coming weeks, you will essentially perform one round of SELEX. Because SELEX typically takes several rounds to isolate target-binding aptamers, you will start with a known aptamer mixture rather than than a completely random library. Your goal will be to explore what experimental parameters affect the enrichment of a heme-binding RNA aptamer from a mixture of heme-binding and non-binding RNAs. | In this module you will investigate RNA aptamer selection. You may already be familiar with peptides or proteins, such as antibodies, that bind to specific molecules. Short fragments of RNA – called RNA aptamers - can have secondary structures that also allow them to bind a target molecule with good affinity and specificity. Normally, RNA aptamers that bind particular targets are found by screening many candidates at random in a process called SELEX, or systematic evolution of ligands by exponential enrichment; however, predictive computational tools can also be used. In the coming weeks, you will essentially perform one round of SELEX. Because SELEX typically takes several rounds to isolate target-binding aptamers, you will start with a known aptamer mixture rather than than a completely random library. Your goal will be to explore what experimental parameters affect the enrichment of a heme-binding RNA aptamer from a mixture of heme-binding and non-binding RNAs. | ||
[ | We thank 20.109 instructor [[Natalie Kuldell]] for helpful discussions and for acquiring funding for module development. | ||
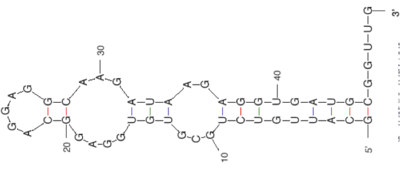
[[Image:20109_6-5_crop.png|thumb|center|400px|'''Structure of a putative aptamer''' Image was prepared using ''RNA folding'' at http://mfold.bioinfo.rpi.edu/]] | |||
[[20.109(S10):Amplify aptamer-encoding DNA (Day1)| Module 1 Day 1: Amplify aptamer-encoding DNA]]<br> | |||
[[20.109(S10):Purify aptamer-encoding DNA (Day2)| Module 1 Day 2: Purify aptamer-encoding DNA]]<br> | |||
[[20.109(S10):Prepare RNA by IVT (Day3)| Module 1 Day 3: Prepare RNA by IVT]]<br> | |||
[[20.109(S10):Purify RNA and run affinity column (Day4)| Module 1 Day 4: Purify RNA and run affinity column]]<br> | |||
[[20.109(S10):RNA to DNA by RT-PCR (Day5)| Module 1 Day 5: RNA to DNA by RT-PCR]]<br> | |||
[[20.109(S10):Post-selection IVT and journal club (Day6)| Module 1 Day 6: Post-selection IVT and journal club]]<br> | |||
[[20.109(S10):Aptamer binding assay (Day7)| Module 1 Day 7: Aptamer binding assay]]<br> | |||
[[20.109(S10):Journal club (Day8)| Module 1 Day 8: Journal club]]<br> | |||
[[20.109(S10): RNA engineering report | Laboratory Report]]<br> | |||
[[20.109(S10): RNA computational analysis | Computational analysis]] | |||
[[20.109(S10): TA notes for module 1| TA notes, mod 1]] | |||
Latest revision as of 10:34, 2 February 2010
Module 1
Instructors: Jacquin Niles and Agi Stachowiak
TA: Christina Birch
In this module you will investigate RNA aptamer selection. You may already be familiar with peptides or proteins, such as antibodies, that bind to specific molecules. Short fragments of RNA – called RNA aptamers - can have secondary structures that also allow them to bind a target molecule with good affinity and specificity. Normally, RNA aptamers that bind particular targets are found by screening many candidates at random in a process called SELEX, or systematic evolution of ligands by exponential enrichment; however, predictive computational tools can also be used. In the coming weeks, you will essentially perform one round of SELEX. Because SELEX typically takes several rounds to isolate target-binding aptamers, you will start with a known aptamer mixture rather than than a completely random library. Your goal will be to explore what experimental parameters affect the enrichment of a heme-binding RNA aptamer from a mixture of heme-binding and non-binding RNAs.
We thank 20.109 instructor Natalie Kuldell for helpful discussions and for acquiring funding for module development.
Module 1 Day 1: Amplify aptamer-encoding DNA
Module 1 Day 2: Purify aptamer-encoding DNA
Module 1 Day 3: Prepare RNA by IVT
Module 1 Day 4: Purify RNA and run affinity column
Module 1 Day 5: RNA to DNA by RT-PCR
Module 1 Day 6: Post-selection IVT and journal club
Module 1 Day 7: Aptamer binding assay
Module 1 Day 8: Journal club
